

Also, a data structure can be accessed through an abstraction that hides the details of the optimized implementation, and this results in looser coupling between components. Another goal is interoperability: by using the same data structures, multiple routines, spread across different packages, can operate on the data without cumbersome conversions.

The notion of ranges can be made explicit in the application programming interface (API), permitting the expression of algorithms in a succinct and readable language that illustrates concepts instead of exposing implementation details. The primary argument for storing ranges in specialized, formal data structures is efficiency, in terms of both implementation and language. Overlap and nearest neighbor detection is fundamental to the annotation of ChIP-seq peaks, estimating expression from RNA-seq data and many other integrative analyses. Coverage calculation is important for detecting regions of enrichment and for producing visual summaries.
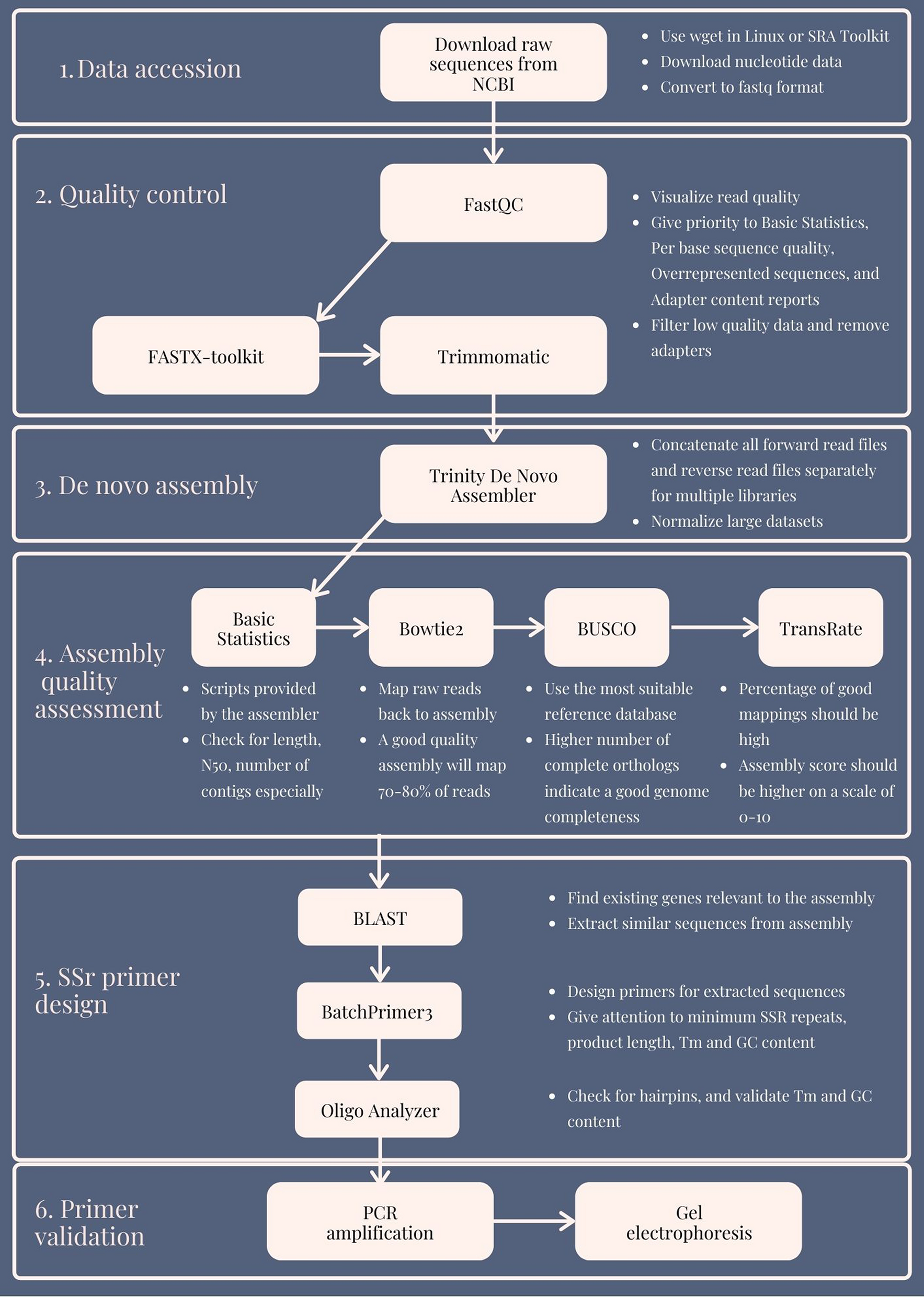
For example, computations on gene models involve set operations on ranges, including intersection, union and complement. Similarly, for RNA-seq data, analysts measure gene expression based on counting the alignments overlapping exons.Īll these analyses depend on specialized, range-based algorithms and data structures. These ranges are then annotated according to their overlap with and proximity to other ranges, such as gene structures. In the analysis of ChIP-seq data, it is typical to calculate the depth of alignment coverage, which then serves as input to calling algorithms which output peaks as ranges. Ranges also play a central role in the analysis of experimental data, where they are used to represent read alignments. Examples include deriving candidate promoter regions, finding introns, calculating the total exonic length of a transcript or finding the exonic regions that are unique to a particular transcript in an alternatively spliced gene. Thus, ranges play a central role in genomic data analysis, and statistical tools should consider ranges to be as fundamental as quantitative and categorical data types.įor example, ranges are integral to the manipulation of gene model annotations. Data integration, within and between those two categories, is made possible by treating the data as ranges on the genome, which acts as a common scaffold. Second, there are primary experimental measurements, such as read alignments from high-throughput sequencing. Such annotations are highly processed and are often served by public databases such as NCBI or EBI.

First, there are the annotations, such as gene models, transcription factor binding site predictions, GC percentage, polymorphisms, and conservation scores. These data fall into two broad categories. The genome is typically represented as a linear sequence, split over multiple chromosomes, and data are linked to the genome by occupying a range of positions on the sequence. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.Ĭompeting interests: The authors have declared that no competing interests exist. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.įunding: This work was funded by the National Institutes of Health, National Human Genome Research Group through grants P41 HG004059 and U41 HG004059 and (for VJC) by National Heart, Lung and Blood Institute grants R01 HL086601, R01 HL093076 and R01 HL094635. Received: JanuAccepted: Published: August 8, 2013Ĭopyright: © 2013 Lawrence et al. PLoS Comput Biol 9(8):Įditor: Andreas Prlic, University of California, San Diego, United States of America (2013) Software for Computing and Annotating Genomic Ranges. Citation: Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, et al.


 0 kommentar(er)
0 kommentar(er)
